Setup GloWPa input directory
devtools::load_all()
## ℹ Loading glowpa
dir.create("glowpa/input/population", showWarnings = FALSE, recursive = TRUE)
dir.create("glowpa/output", showWarnings = FALSE, recursive = TRUE)
Scenario options
gadm_level <- 2
pathogen <- "rotavirus"
Create Raster
vect_gadm <- terra::vect("default/geodata/geodata.shp")
# make numbers
vect_gadm[["iso"]] <- 1:nrow(vect_gadm)
rast_domain <- terra::rast(resolution = .5)
rast_iso <- terra::trim(
terra::rasterize(vect_gadm, rast_domain, field = "iso", touches = TRUE)
)
terra::writeRaster(rast_iso,"glowpa/input/isoraster.tif", overwrite = TRUE)
Create Gridded Population
Note: this should added to the waterpath data service.
rast_pop_urban <- terra::rast("../../../inst/extdata/global/human/pop_urban.tif")
rast_pop_rural <- terra::rast("../../../inst/extdata/global/human/pop_rural.tif")
rast_pop_urban_default <- terra::crop(rast_pop_urban, rast_iso, mask = TRUE)
rast_pop_rural_default <- terra::crop(rast_pop_rural, rast_iso, mask = TRUE)
terra::writeRaster(
rast_pop_urban_default, "glowpa/input/population/pop_urban.tif", overwrite = TRUE)
terra::writeRaster(
rast_pop_rural_default, "glowpa/input/population/pop_rural.tif", overwrite = TRUE)
Create Isodata
df_population <- read.csv("default/population.csv", sep = ",")
df_sanitation <- read.csv("default/sanitation.csv", sep = ",")
df_population %>%
dplyr::mutate_if(is.numeric, format, digits = 3, scientific = TRUE)
| iso | gid | iso3 | subarea | hdi | population | fraction_urban_pop | fraction_pop_under5 |
|---|---|---|---|---|---|---|---|
| GRC.1.1_1 | GRC.1.1_1 | GRC | North Aegean | 8.93e-01 | 179523 | 3.37e-01 | 4.2e-02 |
| GRC.1.2_1 | GRC.1.2_1 | GRC | South Aegean | 8.93e-01 | 303711 | 4.38e-01 | 4.2e-02 |
gid_col <- sprintf("GID_%s", gadm_level)
df_population <- df_population %>% dplyr::rename(iso_country = "iso3")
df_sanitation <- df_sanitation %>%
dplyr::rename(iso_country = "alpha3",
onsiteDumpedland_rur = "onsiteDumpedLand_rur",
onsiteDumpedland_urb = "onsiteDumpedLand_urb")
df_gadm <- as.data.frame(vect_gadm) %>% dplyr::select(iso,dplyr::one_of(gid_col))
join_by = dplyr::join_by(!!sym(gid_col) == iso)
df_isodata <- df_gadm %>%
dplyr::left_join(df_population, by = join_by ) %>%
dplyr::left_join(df_sanitation, by = "iso_country")
saveRDS(df_isodata,"glowpa/input/isodata.RDS")
Create Treatment Data
df_treatment <- read.csv("default/treatment.csv", sep = ",")
df_treatment <- df_treatment %>% dplyr::mutate(
treatment_type = stringr::str_to_title(treatment_type)
)
saveRDS(df_treatment, "glowpa/input/wwtp.RDS")
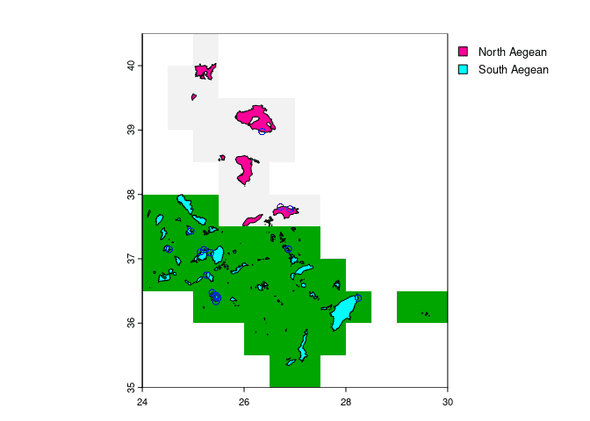
Make Map
vect_treatment <- terra::vect(df_treatment, geom = c("lon", "lat"))
terra::plot(rast_iso, legend = FALSE)
terra::plot(vect_gadm, "NAME_2" ,add = TRUE)
terra::plot(vect_treatment, col = "blue", pch = 1 ,add = TRUE)
Make the configuration file
list_config = list(
input = list(
isoraster = "input/isoraster.tif",
isodata = "input/isodata.RDS",
wwtp = "input/wwtp.RDS",
population = list(
urban = "input/population/pop_urban.tif",
rural = "input/population/pop_rural.tif"
)
),
pathogen = pathogen,
wwtp = list(
treatment = "POINT"
),
logger = list(
enabled = TRUE,
threshold = "INFO",
appender = "CONSOLE"
),
output = list(
dir = "output"
)
)
response = configr::write.config(list_config,"glowpa/default.yaml", write.type = "yaml")
Run GloWPa
wd <- getwd()
setwd("glowpa")
glowpa::glowpa_init("default.yaml")
glowpa::glowpa_start()
setwd(wd)
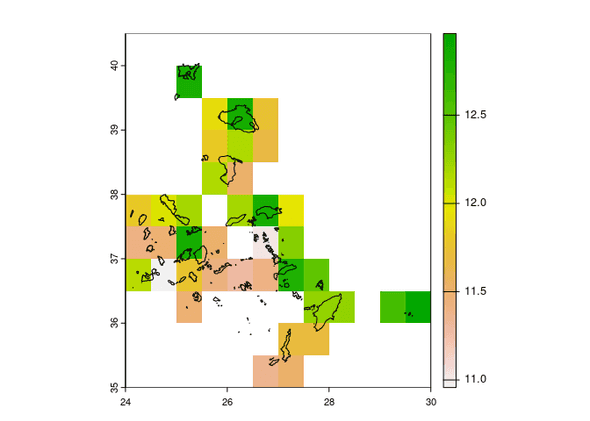
Make Plots
Loads showed in log10 transformation
glowpa_run <- glowpa::glowpa_get_run()
df_human_sources <- read.csv(
file.path("glowpa/output", glowpa_run$settings$output$sources$human$surface_water))
vect_human_sources <- terra::merge(vect_gadm, df_human_sources, by = "iso")
rast_surface_water <- terra::rast(
file.path("glowpa/output", glowpa_run$settings$output$sinks$surface_water$grid))
terra::plot(log10(rast_surface_water))
terra::plot(vect_gadm, add = TRUE)
Loads in Sinks
Loads showed in log10 transformation
df_sinks <- read.csv(
file.path("glowpa/output", glowpa_run$settings$output$sinks$surface_water$table)) %>%
dplyr::mutate_at(
.vars = c("humans", "land","wwtp"), .funs = log10
)
df_sinks
| iso | humans | land | wwtp |
|---|---|---|---|
| 1 | 13.45878 | 8.951446 | 10.70332 |
| 2 | 13.63303 | 9.147552 | 11.87863 |
Human Attribution
Loads showed in log10 transformation
table_human_sources <- df_human_sources %>%
dplyr::mutate_at(.vars = glowpa:::CONSTANTS$SANITATION_TYPES, .funs = log10) %>%
dplyr::mutate_if(is.numeric,format, digits = 4, scientific = TRUE) %>%
dplyr::rename_with(~ stringr::str_trunc(.x, 10))
table_human_sources %>% dplyr::select(iso, 2:8)
| iso | bucketL… | compost… | contain… | flushOpen | flushPit | flushSe… | flushSewer |
|---|---|---|---|---|---|---|---|
| 1 | -Inf | -Inf | -Inf | -Inf | -Inf | 1.151e+01 | 1.345e+01 |
| 2 | -Inf | -Inf | -Inf | -Inf | -Inf | 1.171e+01 | 1.364e+01 |
table_human_sources %>% dplyr::select(iso, 9:14)
| iso | flushUn… | hanging… | openDef… | other | pitNoSlab | pitSlab |
|---|---|---|---|---|---|---|
| 1 | -Inf | -Inf | -Inf | -Inf | -Inf | -Inf |
| 2 | -Inf | -Inf | -Inf | -Inf | -Inf | -Inf |